Exploring lncRNA Expression Patterns in Patients With Hypertrophied Ligamentum Flavum
Article information
Abstract
Objective
Hypertrophy ligamentum flavum (LFH) is a common cause of lumbar spinal stenosis, resulting in significant disability and morbidity. Although long noncoding RNAs (lncRNAs) have been associated with various biological processes and disorders, their involvement in LFH remains not fully understood.
Methods
Human ligamentum flavum samples were analyzed using lncRNA sequencing followed by validation through quantitative real-time polymerase chain reaction. To explore the potential biological functions of differentially expressed lncRNA-associated genes, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed. We also studied the impact of lncRNA PARD3-AS1 on the progression of LFH in vitro.
Results
In the LFH tissues when compared to that in the nonhypertrophic ligamentum flavum (LFN) tissues, a total of 1,091 lncRNAs exhibited differential expression, with 645 upregulated and 446 downregulated. Based on GO analysis, the differentially expressed transcripts primarily participated in metabolic processes, organelles, nuclear lumen, cytoplasm, protein binding, nucleic acid binding, and transcription factor activity. Moreover, KEGG pathway analysis indicated that the differentially expressed lncRNAs were associated with the hippo signaling pathway, nucleotide excision repair, and nuclear factor-kappa B signaling pathway. The expression of PARD3-AS1, RP11-430G17.3, RP1-193H18.3, and H19 was confirmed to be consistent with the sequencing analysis. Inhibition of PARD3-AS1 resulted in the suppression of fibrosis in LFH cells, whereas the overexpression of PARD3-AS1 promoted fibrosis in LFH cells in vitro.
Conclusion
This study identified distinct expression patterns of lncRNAs that are linked to LFH, providing insights into its underlying mechanisms and potential prognostic and therapeutic interventions. Notably, PARD3-AS1 appears to play a significant role in the pathophysiology of LFH.
INTRODUCTION
Lumbar spinal stenosis (LSS) is a prevalent condition, particularly among older adults, characterized by the constriction of the lumbar spinal canal. This constriction can lead to the compression of the spinal cord and/or nerve roots, resulting in pain, sensory changes, and limited mobility [1]. Hypertrophy ligamentum flavum (LFH) plays an important role in the occurrence and development of LSS [2]. LFH involves an increase in the thickness and stiffness of the LF, which is a yellow elastic connective tissue located between adjacent laminae. LFH is characterized by the loss of elastic fibers and the increase of collagen fibers, leading to ligamentum flavum (LF) fibrosis, spinal canal stenosis, and compression of neural elements [2-5]. Actually, ligamentum flavum thickness is positively correlated with the degree of fibrosis, which is the main cause of LFH [2,5-7]. However, the exact mechanisms underlying LFH remain to be fully understood.
Long noncoding RNAs (lncRNAs) are noncoding RNA molecules longer than 200 nucleotides that influence gene expression at various levels, including transcription, post-transcriptional regulation, and epigenetic modulation [8]. The role of lncRNAs has been identified in diverse biological processes, including cell proliferation, apoptosis, and tumorigenesis [9,10]. LncRNAs have also been associated with musculoskeletal diseases, such as osteoarthritis [11], intervertebral disc degeneration [12], and Ossification of the ligamentum flavum [13]. lncRNAs are also involved in the pathological process of fibrosis of the lung [14,15], liver [16,17], kidney [18,19], and heart [20,21]. Past studies in LFH have demonstrated that lncRNAs are significantly associated with LF thickness and fibrosis in LSS patients, such as lncXIST [22]. However, the understanding of lncRNAs in the context of LFH is still limited. The study of differentially expressed lncrnas in LF of LSS patients can facilitate further elucidation of the pathogenesis of LFH at the epigenetic level, which may provide new clues for the treatment of LFH. This study aimed to investigate the differential expression of lncRNAs in LF tissues obtained from individuals with LSS when compared to LFN samples. We employed lncRNA sequencing analysis to identify the altered lncRNAs and validated the findings using quantitative real-time polymerase chain reaction (qRT-PCR) in LF samples. Additionally, we conducted pathway analysis using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) to determine the potential biological functions of the differentially expressed lncRNAs. Furthermore, we examined the impact of lncRNA PARD3-AS1 on the progression of LFH in vitro. Our findings may offer new insights into the mechanisms underlying LFH development and contribute to the development of novel diagnostic and therapeutic strategies for this disorder.
MATERIALS AND METHODS
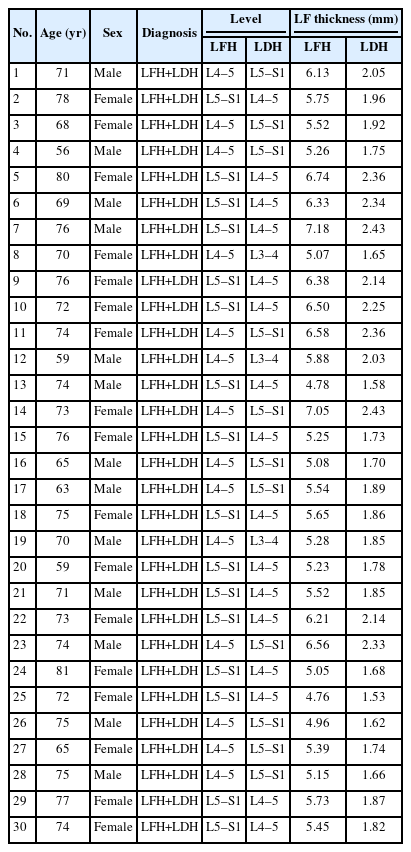
1. Tissue Samples
We screened 30 patients with both LFH and lumbar disc herniation (LDH) from those with LSS symptoms. Clinical diagnosis indicates that both the affected areas of LFH and LDH in these patients require surgical treatment to remove LF. Informed consent was obtained from all patients. For the LFH samples, we determined the location of LFH disease through intraoperative inspection and biopsy and removed them. For the LFN samples, we used LF samples from LDH-affected areas with no evidence of LSS based on preoperative magnetic resonance imaging. The tissues were immediately frozen in liquid nitrogen upon collection and stored at -80°C until further analysis.
The ethics committee of Renji Hospital approved this study, which is consistent with the guidelines of the Declaration of Helsinki. All patients provided their written informed consent prior to study participation.
2. Histological Analyses
The ligamentum flavum tissues were fixed overnight in 4% paraformaldehyde (Beyotime, Shanghai, China), followed by embedding in paraffin using standard procedures. Serial sections with a thickness of 4 μm were prepared for staining with hematoxylin and eosin (HE) and Masson trichrome.
3. RNA Extraction and Sequencing
As previously described, lncRNA sequencing was performed on 3 LFH and LFN samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) [23]. The integrity of the RNA was assessed using NanoDrop ND-1000 (NanoDrop Thermo, Waltham, MA, USA). The Ribo-Zero ribosomal RNA removal kit (Illumina, San Diego, CA, USA) was used to remove ribosomal RNA, and strand-specific total RNA libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit (Illumina). Sequencing was performed by Cloud-Seq Biotech (Shanghai, China) on an Illumina HiSeq 2500 Sequencer (Illumina).
4. Bioinformatics Analysis
The high-quality reads were aligned to the human reference genome (UCSC hg19) using Hisat2 software (ver. 2.0.4). Fragments per kilobase of transcript per million mapped reads (FPKM) values for lncRNAs were obtained using the cuffdiff software (part of cufflinks). Fold change and p-values were calculated based on FPKM, and differentially expressed lncRNAs were identified using a volcano plot and fold-change filters. Hierarchical clustering was performed using the differentially expressed lncRNA data. GO enrichment and KEGG pathway analyses were conducted to explore the potential biological functions of the differentially expressed lncRNAs using GO and KOBAS software (the KEGG orthology-based annotation system).
5. Isolation of Primary LF Cells
LF cells were isolated following a previously described method. The LFH and LFN tissues were minced into small pieces measuring approximately 0.5–1 mm3 and treated with 0.25% trypsin and 250 U/mL type I collagenase (Sigma-Aldrich, St. Louis, MO, USA) in T25 flasks. The resulting cells were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco, Waltham, MA, USA) at 37°C in a humidified incubator with 5% CO2 until they attained confluency. The culture media was replenished every 3 days, and trypsin was used for cell detachment during passaging. LF cells obtained from the third passage were used for all experiments.
6. Plasmids and Transduction
To investigate the role of lncRNA PARD3-AS1 in LFH progression, in vitro experiments were conducted using human LFH cells obtained from the tissue samples. shRNAs targeting sequences shPARD3-AS1-1 (5´-CCTAACTCGGTCCACGTTCC-3´), shPARD3-AS1-2 (5´-GTTCCCTAACTCGGTCCACG-3´), shPARD3-AS1-3 (5´-CGTGGACCGAGTTAGGGAAC-3´), and PARD3-AS1 overexpressing plasmid (ex-PARD3-AS1) were purchased from Vigenebio (Shandong, China). LFH cells were transfected with shRNA sequences/negative control or overexpressing plasmid/empty vector (mock) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was assessed using qRT-PCR at 48-hour posttransfection.
7. Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from LF tissues or cells using TRIzol reagents (Invitrogen), followed by reverse transcription. qPCR was performed on the Applied Biosystems 7300 Fast Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) using the SYBR Green qPCR master mix (Qiagen, Shanghai, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. RNA expression was evaluated by the 2−ΔΔCT method [24]. Sequences of the primers used are listed in Table 1.
8. Western Blotting
Total cellular protein was extracted using the radioimmunoprecipitation lysis buffer with protease inhibitors (Beyotime). Protein levels in each sample were quantified using a bicinchoninic acid assay kit (Beyotime). These proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and 40 μg of the proteins were subsequently transferred to polyvinylidene fluoride membranes.
The blots were blocked with 5% skim milk and incubated overnight at 4°C with primary antibodies. These antibodies (Abcam, Cambridge, UK) were prepared in 5% bovine serum albumin as follows: collagen I (1:1,500, ab279711); collagen III (1:1,500, ab6310); transforming growth factor (TGF)-β1 (1:1,500, ab215715), and GAPDH (1:5,000, ab8245) [25-28].
After incubation with suitable horseradish peroxidase-conjugated secondary antibodies for 1 hour, protein bands were visualized using an enhanced chemiluminescence kit (Millipore, Darmstadt, Germany). Densitometric quantification was performed using ImageJ (National Institutes of Health, Bethesda, MD, USA), with GAPDH serving as a normalization control.
9. Statistical Analysis
The study data were analyzed using GraphPad Prism v8 (GraphPad Software, San Diego, CA, USA) and presented as the means± standard deviation of 3 independent experiments. Differences in the groups were analyzed by Student t-tests or 1-way analysis of variance. A p-value of < 0.05 was considered statistically significant.
RESULTS
1. Differential Expression of lncRNAs in LFH and LFN Tissues
lncRNA expression patterns in patients with LFH were investigated by performing RNA sequencing (RNA-seq) on LFH tissue samples (n = 3) and compared with those of control LFN samples (n = 3). The histological examination of patient-derived LF specimens stained with HE showed disrupted fiber organization in the LFH tissues (Fig. 1A). Furthermore, Masson trichrome staining highlighted the increased abundance of collagen fibers in these LFH tissues (Fig. 1B). The RNA-seq analysis revealed that 1,091 lncRNAs exhibited differential expression between LFH and LFN tissues, and 645 upregulated and 446 downregulated lncRNAs were observed in LFH tissues (Fig. 1C; Supplementary Data 1). The top 10 upregulated and downregulated lncRNAs on the basis of fold change values are displayed in a heat map (Fig. 1D) and summarized in Table 2.
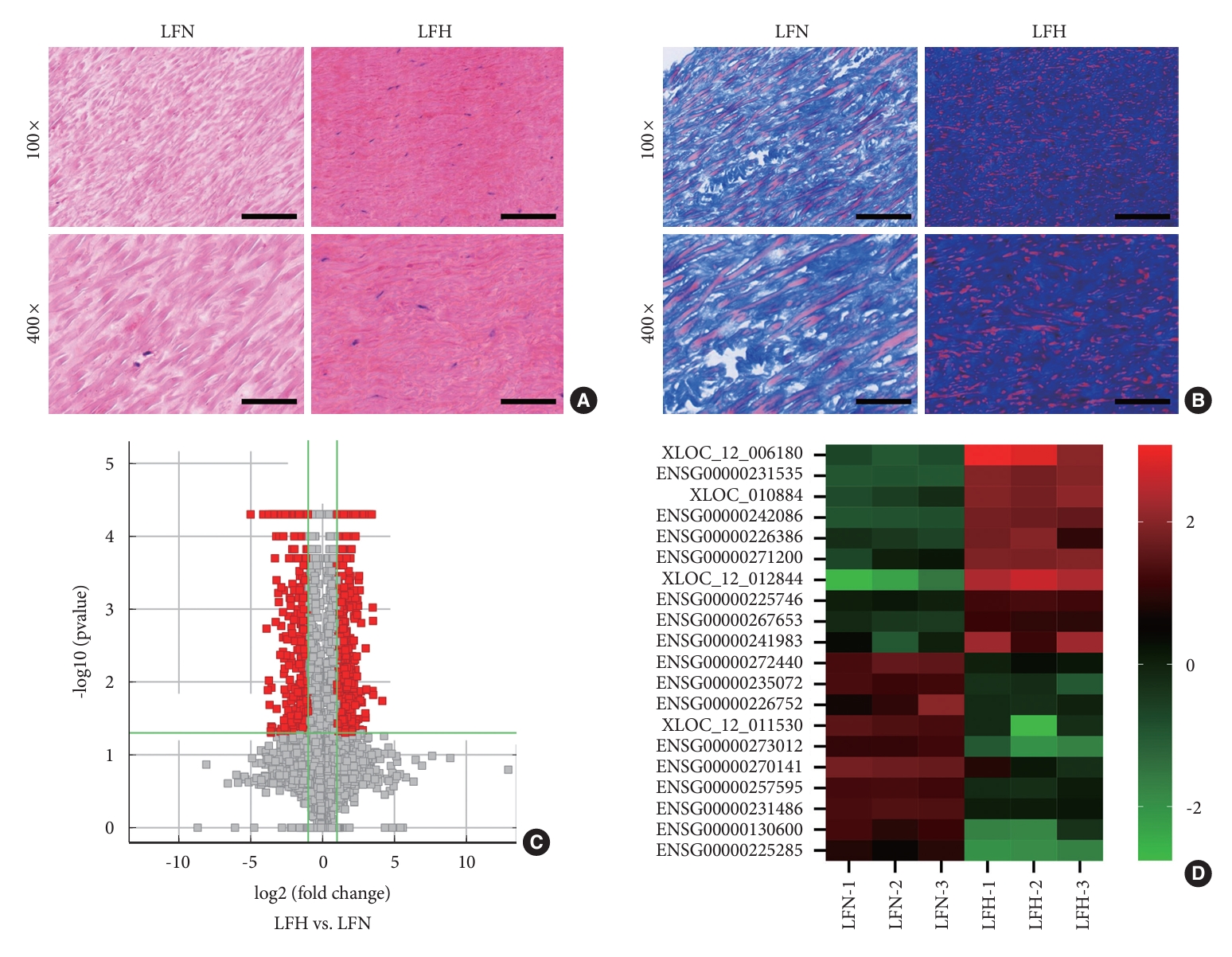
Differences in the lncRNA expression profiles between LFH and LFN tissues. (A) Illustrative images depicting the results of hematoxylin and eosin staining. (B) Exemplary images of the results of Masson’s trichrome staining. Scale bar=100 µm or 20 µm. (C) Volcano plot displaying the RNA-seq data. (D) Heat maps depictng the varying expressions of distinct long noncoding RNAs in both LFN and LFH tissues. LFH, hypertrophy ligamentum flavum; LFN, nonhypertrophic ligamentum flavum.
2. Functional Annotation of the Differentially Expressed Lncrnas
GO enrichment and KEGG pathway analyses were performed to identify the potential biological functions of the differentially expressed lncRNAs. The top 10 most-enriched GO terms were associated with biological processes, cellular components, and molecular functions (Fig. 2) as follows: The differentially expressed transcripts were mainly involved in metabolic processes of the biological process domain (Fig. 2A and D); the organelles, nuclear lumen, and cytoplasm of the cellular component domain (Fig. 2B and E); and protein binding, nucleic acid binding, and transcription factor activity of the molecular function domain (Fig. 2C and F).
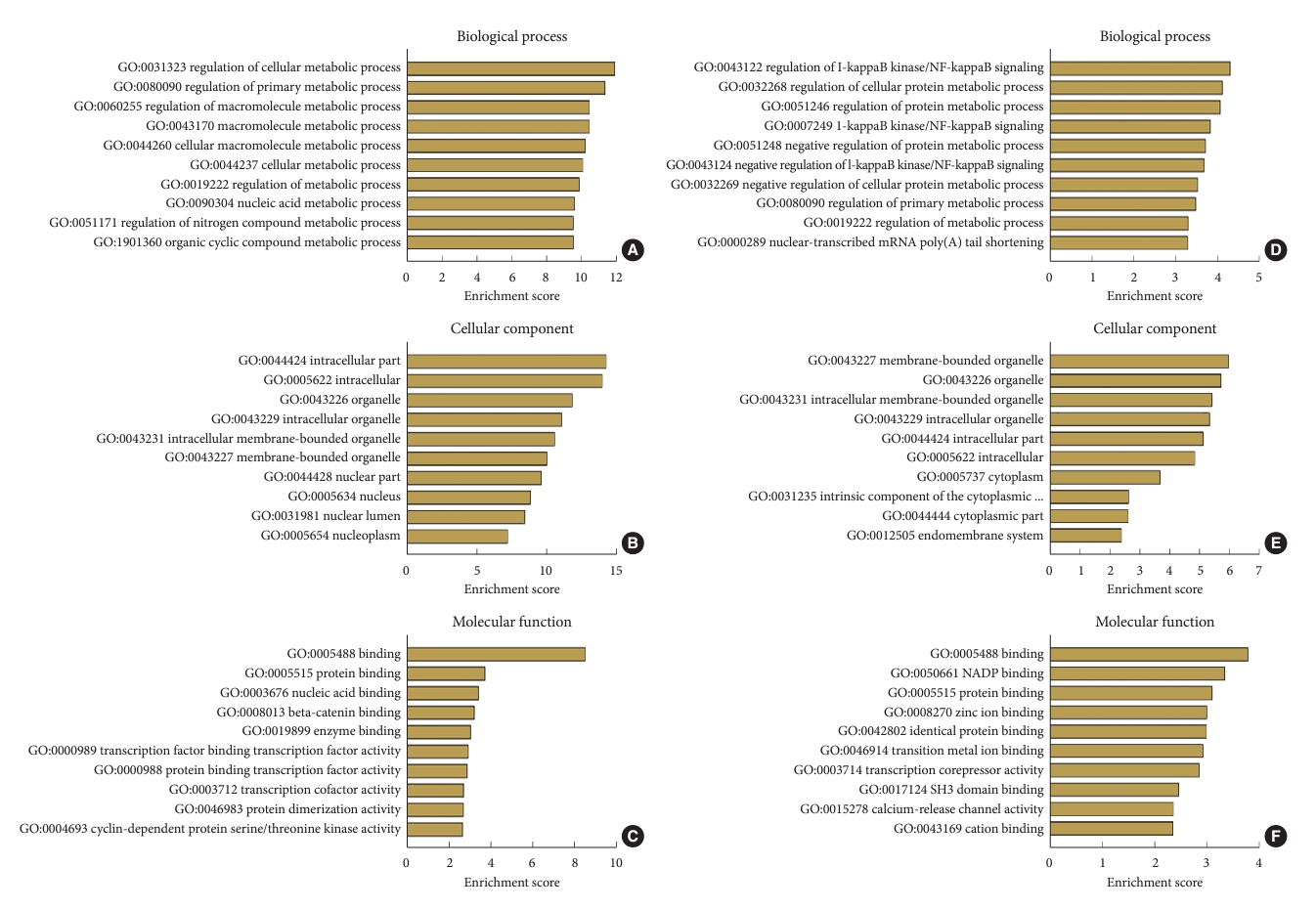
Gene Ontology (GO) enrichment analysis for differentially expressed long noncoding RNAs (lncRNAs). (A–C) The top 10 enrichment score values for the significant GO enrichment terms in the biological process (A), cellular component (B), and molecular function (C) categories for upregulated lncRNAs. (D–F) The top 10 enrichment score values for the significant GO enrichment terms in the biological process (D), cellular component (E), and molecular function (F) categories for downregulated lncRNAs.
The top 10 and 5 enriched KEGG pathways of upregulated and downregulated circular RNAs (circRNAs), respectively (Fig. 3), include the hippo signaling pathway and nucleotide excision repair for upregulated circRNAs (Fig. 3A) and SNARE interactions in vesicular transport and the nuclear factor (NF)-kappa B signaling pathway for downregulated circRNAs (Fig. 3B).

The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for differentially expressed long noncoding RNAs (lncRNAs). (A) The top 10 enrichment score values for the significantly enriched pathways in upregulated lncRNAs. (B) The top 10 enrichment score values for the significantly enriched pathways in downregulated lncRNAs.
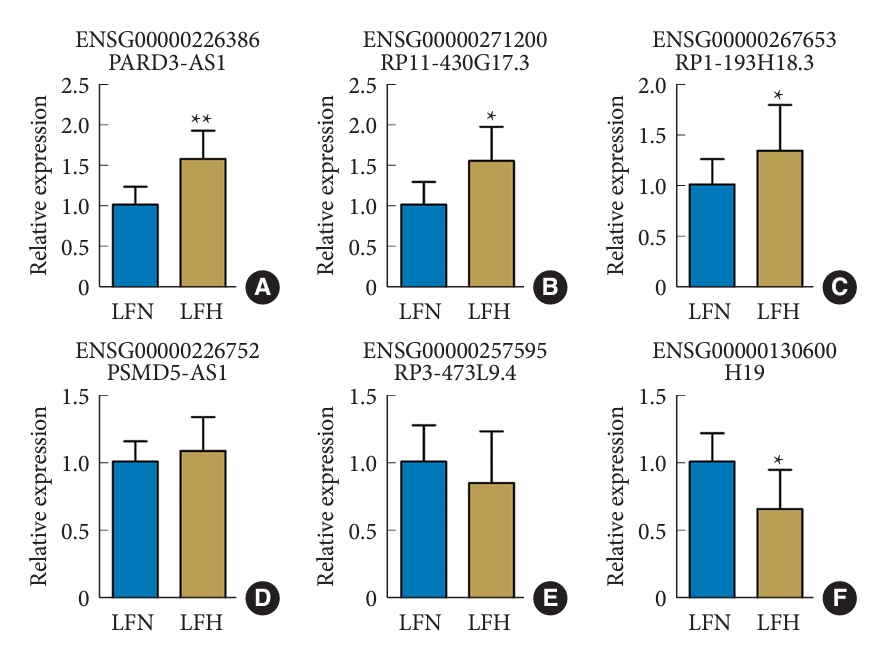
3. Validation of the Altered LncRNAs by qRT-PCR
To validate the differential expression of the altered lncRNAs identified by the sequencing analysis, we performed qRT-PCR on 30 paired tissue samples. The clinical and radiographic data are shown in Table 3. The upregulated and downregulated lncRNAs were arranged according to the fold change from high to low. Three randomly selected upregulated and downregulated lncRNAs from among the top 10 lncRNAs were arranged, namely PARD3-AS1 (Fig. 4A), RP11-430G17.3 (Fig. 4B), RP1-193H18.3 (Fig. 4C), PSMD5-AS1 (Fig. 4D), RP3-473L9.4 (Fig. 4E), and H19 (Fig. 4F), for validation. The qRT-PCR results showed that the expression patterns of PARD3-AS1, RP11-430G17.3, RP1-193H18.3, and H19 were consistent with the sequencing analysis results, which confirmed the differential expression of these lncRNAs. Among the 6 lncRNAs, PARD3-AS1 displayed the largest significant difference.
4. Effect of lncRNA PARD3-AS1 on LFH Progression In Vitro
To investigate the role of lncRNA PARD3-AS1 in LFH progression, we performed in vitro experiments using human LFH cells obtained from the tissue samples. First, we confirmed the transfection efficiency of the knockdown (Fig. 5A) and overexpression (Fig. 5B) of PARD3-AS1 by qRT-PCR. Among the tested shRNA constructs, shPARD3-AS1-3 produced the highest knockdown efficiency and was thus used in downstream experiments. The western blotting results showed that PARD3-AS1 knockdown decreased collagen I, collagen III, and TGF-β1 levels in the LFH cells compared with those in the control cells, while the levels were close to those in LFN cells. However, the PARD3-AS1 overexpression increased collagen I, collagen III, and TGF-β1 levels (Fig. 5C-F).
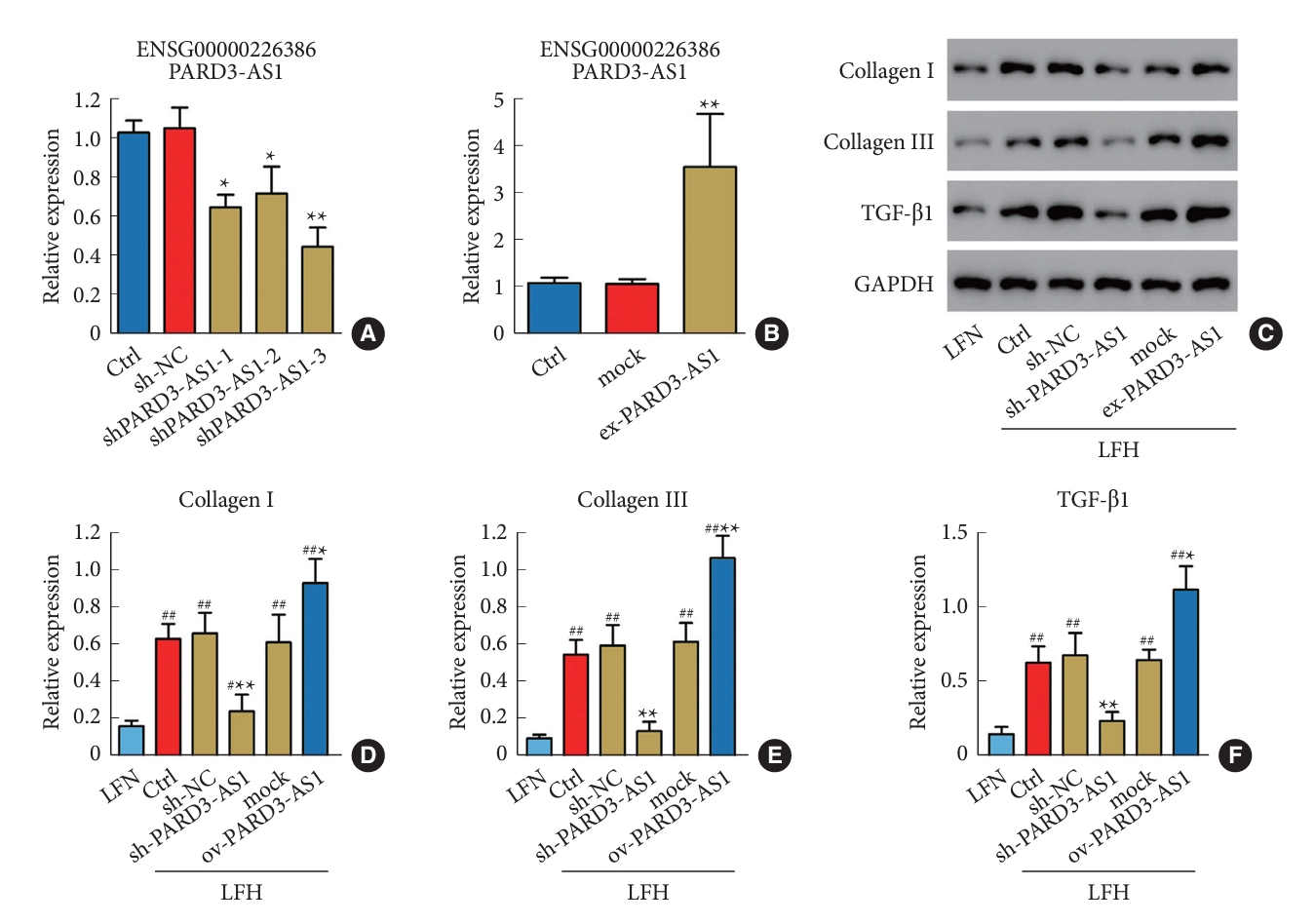
Inhibition of PARD3-AS1 reduces fibrosis in ligamentum flavum cells derived from the LFH tissues. Quantitative real-time polymerase chain reaction was performed to analyze the PARD3-AS1 knockdown (A) and overexpression (B). (C–F) The expression of collagen I, collagen III, and TGF-β1 was evaluated through Western blotting. A significant difference was observed in comparison to control cells (Ctrl), with *p<0.05 and **p<0.01. A significant difference was observed in comparison to LFN cells, with #p<0.05 and ##p<0.01. LFH, hypertrophy ligamentum flavum; LFN, nonhypertrophic ligamentum flavum; TGF, transforming growth factor; sh-NC, shRNA sequences/negative control.
DISCUSSION
LFH is a major cause of LSS, leading to remarkable disability and morbidity [2]. Growing evidence shows that noncoding RNAs, including lncRNAs, play a role in the development and progression of various disorders, including spinal diseases [9,29]. Previous studies have investigated molecular mechanisms underlying LFH; however, the role of lncRNAs in LFH progression is only partially understood [22,23,30,31]. Here, we investigated the expression patterns of lncRNAs in LFH and LFN tissues by lncRNA sequencing. A total of 1,091 differentially expressed lncRNAs were identified in the LFH and LFN tissues, of which 645 were upregulated and 446 were downregulated.
The differentially expressed lncRNAs were analyzed for their biological functions by performing GO enrichment and KEGG pathway analyses. The GO analysis revealed that these lncRNAs were mainly associated with metabolic processes, organelles, nuclear lumen, cytoplasm, protein binding, nucleic acid binding, and transcription factor activity. These results are consistent with those of previous studies that have implicated alterations in cellular metabolism [32], organelle function [33], and transcriptional regulation [34,35] in LFH pathogenesis. Furthermore, the KEGG pathway analysis showed several signaling pathways that might be associated with the differentially expressed lncRNAs, including the hippo signaling pathway, nucleotide excision repair, SNARE interactions in vesicular transport, and the NF-kappa B signaling pathway. The hippo signaling pathway plays a critical role in tissue growth and regeneration [36-38], whereas nucleotide excision repair is involved in DNA damage repair [39,40]. SNARE interactions in vesicular transport are related to synaptic transmission and neurodegenerative diseases [41-43], whereas the NF-kappa B signaling pathway is associated with inflammation and immune responses [44,45]. However, the specific roles of these pathways in LFH warrant further investigation.
The qRT-PCR results confirmed the expression patterns of PARD3-AS1, RP11-430G17.3, RP1-193H18.3, and H19, which were consistent with the sequencing analysis results. The investigation of the effect of lncRNA PARD3-AS1 knockdown/overexpression on the progression of LFH in vitro showed that PARD3-AS1 knockdown decreased the levels of fibrosis-related proteins (collagen I, collagen III, and TGF-β1) in the LFH cells, whereas PARD3-AS1 overexpression increased the levels these proteins. Past studies have demonstrated that collagen I and collagen III are representative fibrotic products [25]. TGF-β promotes collagen production and induces fibrosis in LF cells [26-28]. These results suggest that PARD3-AS1 may significantly affect the underlying processes and pathophysiology of LFH.
Past data has demonstrated that PARD3-AS1 is highly upregulated in the blood vessels of patients with atherosclerotic plaque formation (APF) [46]. PARD3-AS1 is one of the unique genes associated with arterial spasm due to reduced blood flow [47]. Past studies have indicated that vascular disease is one of the contributing factors to spinal degenerative diseases [48]. Patients with LSS are often complicated by peripheral artery diseases (PADs) and aortic diseases based on atherosclerosis (AS) [49]. APF can reduce the blood supply to the lumbar spine and its surrounding structures, hinder the nutrition acquisition of the intervertebral disc, and induce disc degeneration and pain [50]. It is thus speculated that PARD3-AS1 may have a regulatory role between APF and LSS, and PARD3-AS1 is expected to provide a therapeutic target for spinal degenerative diseases induced by vascular diseases. Exploring the mechanism of PARD3-AS1 involvement in LSS can facilitate the elucidation of the pathogenesis of angiomatoid fibrous histiocytoma from the perspective of vascular diseases. The present findings suggest that PARD3-AS1 can promote the synthesis of fibrosis-related proteins in LF and thereby provide a therapeutic clue for LSS patients complicated with vascular diseases, such as APF and PAD. In the future, exploring the mechanism of PARD3-AS1 regulating LFH and its relationship with blood diseases can be used as a new research direction.
CONCLUSION
In this study, we identified specific lncRNA expression patterns associated with enlarged LF in patients with LSS. The differentially expressed lncRNAs might be associated with LFH development via diverse biological processes and pathways, offering a basis for potential prognostic and treatment options for the patients. PARD3-AS1 might have a significant effect on the underlying processes and pathophysiology of LFH and needs further investigation. However, the present study has some limitations, such as a small sample size and the lack of functional experiments in vivo. Thus, studies with larger sample sizes and animal models are warranted to verify our findings and explore the specific roles of altered lncRNAs and genes in LFH pathogenesis.
Supplementary Material
Supplementary Data 1 can be found via https://doi.org/10.14245/ns.2346994.497.
Notes
Conflict of Interest
The authors have nothing to disclose.
Funding/Support
The study was supported by the Shanghai Baoshan District Science and Technology Innovation Special Fund Project (No: 20-E-33), Scientific Research Project of Shanghai Municipal Health Commission (No. 202040370) and Clinical Research Innovation Cultivation Fund of Baoshan Branch of Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (2023-rbcxjj-004).
Author Contribution
Conceptualization: Junling C, GZ, Jianwei C; Data curation: Junling C, MQ, WK, JX; Formal analysis: MQ, WK, JX; Funding acquisition: GZ; Methodology: Junling C, WK, JX, Jianwei C; Project administration: Junling C, GZ, Jianwei C; Writing - original draft: Junling C, MQ, JX; Writing - review & editing: Jianwei C.